Whether in the context of febrile illness, mild delirium, or the dreaded “weak and dizzy,” sepsis lurks around every corner. Then, in an era replete with serious respiratory viruses such as SARS-CoV-2, influenza, and respiratory syncytial virus, the challenge persists of differentiating systemic viral illness from bacteremia. However, where practicing clinicians see problems, diagnostics companies see opportunities.
Explore This Issue
ACEP Now: Vol 41 – No 07 – July 2022Diagnostic and Supplemental Testing
Most emergency physicians are well acquainted with the process of teasing out a diagnosis of infection from otherwise deranged physiology, and likewise further clarifying an underlying bacterial source. As the pressures mount for ever-earlier intervention and even greater diagnostic accuracy, clinical evaluation is supplemented by laboratory testing. Generationally, the simplest tool remains the complete blood count (CBC) and differential, using the white blood cell count and its differentiation between neutrophils, lymphocytes, and other immature forms as clues to further inform the presence and type of infection.
The well-described limitations in sensitivity and specificity for the CBC have led further afield to supplementary tests. Most commonly, and dependent upon local practice patterns, these supplemental tests are typically C-reactive protein (CRP) and procalcitonin. These non-specific markers of systemic inflammation provide incremental predictive value in determining the presence of a serious bacterial infection. Unfortunately, each of these tests generally displays a normal result in concert with a correspondingly benign clinical picture, and a grossly abnormal result when infection is clearly present. In cases where a diagnosis is less clear, results from these tests tend to land squarely in uninformative, indeterminate ranges. Furthermore, each test may be confounded by chronic inflammatory conditions, or falsely reassuring in immunosuppressed patients and early in a disease process.
Despite the marketing push behind procalcitonin over the past decade, growing recognition of its limitations has led to the development of several novel objective tools attempting to improve upon the current state of disarray. One of these is the monocyte distribution width (MDW), branded by Beckman Coulter as the Early Sepsis Indicator (ESId).1 Similar to technologies in automated analyzers in which leukocyte type and red cell size can be evaluated, MDW can likewise be observed. Because monocytes with inflammatory phenotypes increase in size, and these changes may be observed in response to sepsis, MDW has been proposed as another early marker of sepsis.
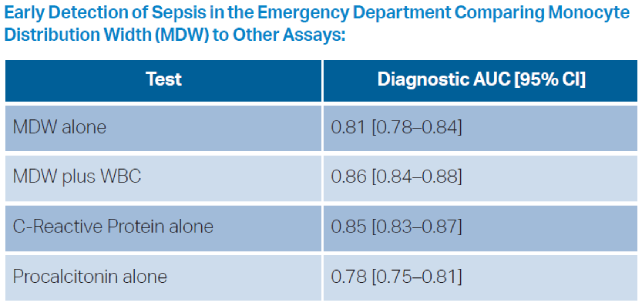
This novel measurement has ultimately shown little added value over current nonspecific markers. Across various studies evaluating its performance, the area under the receiver operating curve (AUROC) for MDW is in the range of 0.70 to 0.80.2 While this has better diagnostic precision than a coin flip, in various retrospective and prospective evaluations MDW performed similarly to both CRP and procalcitonin.3 At the MDW cut-off value of 20 (defined at regulatory approval), sensitivity is reported as 95.5 percent, with a specificity of 26.5 percent. This product thus slots in precariously as a one-way decision tool to reinforce a clinical decision of the absence of sepsis, but with extremely poor positive predictive value.4 The primary advantage of this test compared to CRP or procalcitonin, is that the result is embedded in the CBC, rather than necessitating a separate assay.
Additional proprietary biomarker assays under development, include the IntelliSep and Immunix tests. The IntelliSep test pushes samples through microfluidic channels with a camera performing image acquisition of WBCs under deformation stress.5 The behavior of WBCs under deformation stress is measured by automated methods as their primary metric, reflecting host response to infection. These properties were then correlated with clinical outcomes, as validated on a set of emergency department patients presenting with potential sepsis and a set of healthy volunteers. Similarly to other inflammatory markers, the output of the test is risk-stratification into low- and high-risk cohorts alongside an indeterminate zone.
What the Studies Show
Few published studies of the IntelliSep test exist, and none include direct comparisons against other conventional markers of inflammation.6,7 Using the most conservative interpretation of published performance, the lowest-risk category demonstrated an 87.5 percent sensitivity, while the highest-risk category demonstrated an 86.2 percent specificity. The number analyzed was low enough that even small changes in sepsis outcome adjudication had dramatic effects on positive and negative likelihood ratios. To put it mildly, many data remain to be presented to evaluate both this test’s performance and its feasibility in clinical deployment.
The Immunix test is another biomarkerbased evaluation with a slightly different twist. In this instance, biomarker data is combined with electronic health record (EHR) data to produce a prediction superior to either biomarkers or EHR data in isolation.8,9 For emergency department applications, their proprietary implementation utilizes IL-6, CRP, and procalcitonin, while a hospital-wide version includes additional biomarkers and an expanded set of EHR variables. An even greater paucity of data is available to evaluate this technology for use in the emergency department. A single observational study on frozen remnant blood samples revealed an AUROC on their validation set of 0.83, with a sensitivity of 80 percent and a specificity of 70 percent at their selected optimal threshold to identify a low-risk population.10 In my opinion, this test is even further from operational consideration as it requires the added complexity of direct access to clinical information systems.
The final new test worth discussion is a biomarker marketed to differentiate bacterial and viral infections. The MeMed BV test combines CRP, tumour necrosis factor-related apoptosis-inducing ligand (TRAIL), and interferon-gamma-induced protein-10 (IP-10). Each of these biomarkers in isolation generates an AUROC around 0.60 to 0.68, and their combined characteristics are used to generate a score.11 The scores are then binned into five levels of likelihood of bacterial infection. At the extremes, the positive likelihood ratio is approximately 8.1, with a negative likelihood ratio of 0.1. However, the vast majority of scores have much lower LRs. The U.S. Food and Drug Administration approved this assay based on assessment of equivalence to procalcitonin, leading to the obvious follow-up question of whether it improves on this generally ubiquitous test.12
Generally speaking, all of these potential tools fall far short of demonstrating their value. Each shows some predictive power to address their problem of interest. Many lack prospective, operational comparisons with presently available objective adjuncts to clinical judgment, such as procalcitonin and CRP, and none robustly demonstrate superiority. Most importantly, no studies of these tests report on prospective implementation and impact on either surrogates for sepsis management, such as antibiotic administration and appropriateness, or patient-oriented outcomes relating to morbidity or mortality. While these are exciting times to witness the development of new tools, remain cautious about adoption before their value is proven.
Dr. Radecki is an emergency physician and informatician with Christchurch Hospital in Christchurch, New Zealand. He is the Annals of Emergency Medicine podcast co-host and Journal Club editor and can be found on Twitter @emlitofnote.
References
- Crouser ED, Parrillo JE, Seymour CW, et al. Monocyte distribution width: a novel indicator of sepsis-2 and sepsis-3 in high-risk emergency department patients. Crit Care Med. 2019;47(8):1018–1025.
- Piva E, Zuin J, Pelloso M, et al. Monocyte distribution width (Mdw) parameter as a sepsis indicator in intensive care units. Clin Chem Lab Med. 2021;59(7):1307–1314.
- Woo A la, Oh DK, Park CJ, Hong SB. Monocyte distribution width compared with C-reactive protein and procalcitonin for early sepsis detection in the emergency department. PLoS One. 2021;16(4):e0250101.
- Crouser ED, Parrillo JE, Martin GS, et al. Monocyte distribution width enhances early sepsis detection in the emergency department beyond SIRS and qSOFA. J Intensive Care. 2020;8(1):33.
- Guillou L, Sheybani R, Jensen AE, et al. Development and validation of a cellular host response test as an early diagnostic for sepsis. PLoS One. 2021;16(4):e0246980.
- O’Neal HR, Sheybani R, Caffery TS, et al. Assessment of a cellular host response test as a sepsis diagnostic for those with suspected infection in the emergency department. Crit Care Explor. 2021;3(6):e0460.
- O’Neal HR, Sheybani R, Caffery TS, et al. Assessment of a cellular host response test to risk-stratify suspected COVID-19 patients in the Emergency Department setting. PLoS One. 2022;17(3):e0264220.
- Hassan U, Ghonge T, Reddy B, et al. A point-of-care microfluidic biochip for quantification of CD64 expression from whole blood for sepsis stratification. Nat Commun. 2017;8(1):15949.
- Taneja I, Reddy B, Damhorst G, et al. Combining biomarkers with emr data to identify patients in different phases of sepsis. Sci Rep. 2017;7(1):10800.
- Taneja I, Damhorst GL, Lopez-Espina C, et al. Diagnostic and prognostic capabilities of a biomarker and EMR-based machine learning algorithm for sepsis. Clin Transl Sci. 2021;14(4):1578–1589.
- van der Does Y, Rood PPM, Ramakers C, et al. Identifying patients with bacterial infections using a combination of C-reactive protein, procalcitonin, TRAIL, and IP-10 in the emergency department: a prospective observational cohort study. Clinical Microbiology and Infection. 2018;24(12):1297–1304.
- 510(k) Substantial Equivalence Determination Decision Summary [regulatory government]. United States Food and Drug Administration. 1 Sept 2021. Available at: https://www.accessdata.fda.gov/cdrh_docs/reviews/K210254.pdf. Accessed June 13, 2022.
Pages: 1 2 3 4 | Multi-Page






No Responses to “Diagnosing Sepsis, the Next Generation”